Research in the laboratory
2020 - Current
At the moment, my major research goals involve working with The Wolverine Foundation to advance research and discover novel therapeutic approaches to treat the neuro-developmental disease caused by genetic variations in the gene MAPK8IP3. To accomplish this I am developing patient-guided precision animal zebrafish models, specific to patient variants, using gene editing approaches. I am also developing transgenic lines to assist in visualizing expression of Mapk8ip3 in vivo and completing over expression studies to attempt rescue of phenotypes in a Mapk8ip3 loss-of-function/knock out line. The major goal of this work is to collaborate with other members of The Wolverine Foundation (including the Yeo lab at UAB) and UAB's Center for Precision Animal Modeling team (Yoder lab) to screen drugs in C. elegans, induced pluriopotent stems cells from patients, and in zebrafish patient models to discover drugs that could be used in clinical trials.
2018 - Current
Linking genotype to phenotype: developing patient-guided zebrafish models to precisely identify and treat rare-genetic disorders
As a senior scientist at The Hugh Kaul Precision Medicine Institute (PMI), I work with a team of researchers and physicians to predict the molecular impacts of patient genetic variants and hypothesize treatment options using bioinformatics and artificial intelligence tools, such as our in-house logic-based reasoning program mediKanren. Our goal is to identify readily available, FDA-approved therapeutic options that can be repurposed to treat patients with rare- or ultra-rare genetic disorders for which there is no standard of care. In cases where protein function is unclear, genetic impact is unknown, or few treatment options are predicted, I develop zebrafish models with patient-specific genetic variants or loss-of-function mutations using CRISPR/Cas9 mutagenesis. Zebrafish are excellent research models because they have a sequenced genome, a large research community, and a plethora of established research protocols and tools, including transgenic lines (gene reporters).
2016 - 2018
Understanding fertility: the role of hormone receptors in sex determination, gonad development and gametogenesis
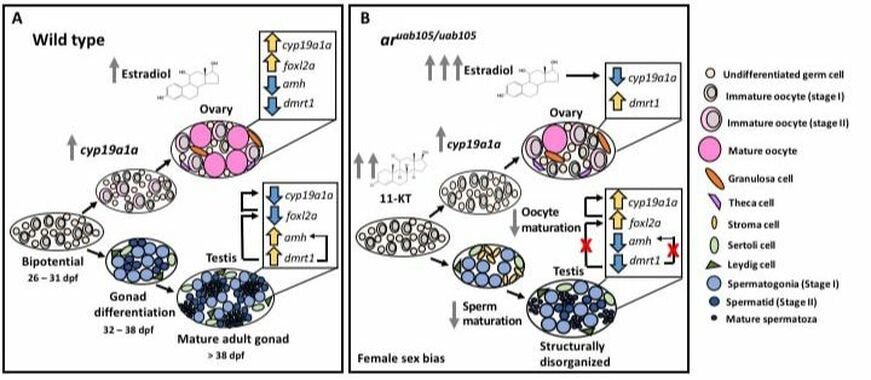
During my postdoc at University of Alabama at Birmingham I utilized zebrafish (Danio rerio) as a model system to study endocrine disruption and the influence of hormones on sex determination and gonad development. Zebrafish are unique in that they lack sex chromosomes and instead rely on a network of genes to determine sexual fate. However, they are extremely sensitive to hormones in their environment and can experience sex bias in their populations when exposed to androgens or estrogens early in their development. Utilizing CRISPR-Cas9 mediated mutagenesis technology, I was able to show that the nuclear androgen receptor (ar), the gene to which testosterone binds, influences sex determination, secondary-sexual characteristics, oocyte maturation and male fertility in zebrafish (Figure 1) (Crowder et al. 2017).
In order to continue exploring how androgen signaling influences sex determination and gonad function, I plan to examine membrane androgen receptor (zip9 and GPCR6A) signaling. Utilizing the aruab105/105 and using CRISPR-Cas9 gene editing I plan to knock out these receptors individually and together with the nuclear androgen receptor to determine the role of each receptor in sex determination, testes organization and spermatogenesis. I would also like to explore the influence of endocrine disrupting chemicals on androgen signaling to determine the mechanism of action of androgen receptor sensitive environmental pollutants and the impact they have on fertility and development. Lastly, I plan to extend this project further to examine how primordial germ-cell fate and differentiation is impacted in mutants lacking normal androgen signaling capabilities.
Figure 1. Proposed mechanisms of how an androgen receptor mutant (uab105/uab105) alters development and gene expression associated with sex determination and gonad maintenance, based on qPCR and histological data. (A) Wild-type zebrafish gonad development is characterized by normal oocyte maturation with equal numbers of stage I, II and III oocytes, organized testes and opposing levels of sex-related genes (arrows represent increased and decreased levels of gene expression relative to controls). (B) AR mutant zebrafish show elevated levels of 11-ketotestosterone and estradiol, leading to decreased oocyte maturation and disorganized testis structure and expression levels of sex-related genes are reversed in adults gonads, more similar to wild-type expression of the opposite sex. Key terms: dpf, days post fertilization; Cyp19a1a, gene coding for the aromatase enzyme that converts androgens to estrogens; Foxl2, Forhead box L2 gene; Amh, anti-mullerian hormone gene; Dmrt1, doublesex and mab-3 related transcription factor.
Research in the classroom
2019
Developing zebrafish models of patient-guided rare-neuropsychiatric disorders
Patients with rare-genetic disorders often exhibit epilepsy-like symptoms, such as seizure and developmental delay. This semester in the Science and Technology honors course: Research Approaches: Molecular Genetics, I am working with students to develop loss-of-function (LOF) zebrafish models for patients with predicted LOF or haploinsufficient genetic variants. Using CRISPR-Cas9 gene editing, students are working on teams of 2 to design CRISPR guides targeting genes of interest, screen RNA guide + Cas9 cutting efficiencies in vivo, and analyze early-stage phenotypes in microinjected F0 zebrafish. Successfully mutated zebrafish will be raised, sequenced, and bred to develop LOF zebrafish models of genetic disorders. Students interested in completing their honor's thesis projects using these LOF mutants will then work with me, in collaboration with Dr. Summer Thyme, to identify screenable phenotypes (behavior, brain structure & activity) for which FDA-approved drug libraries can be screened in order to find therapeutic options to potentially treat individual patients.
2018
Inducing mutations in genes involved in the RAS/MAPK signaling pathway to examine disease mechanisms associated with rare rasopathies
The RAS/MAPK signaling pathway is involved in multiple processes associated with development in humans. Mutations in genes of this pathway can cause a break down in cell signaling necessary for proper growth and development, resulting in a spectrum of rare disorders, collectively referred to as rasopathies. I've partnered with Dr. Matt Might, director of UAB's Hugh Kaul Precison Medicine Institute, to target genes in the RAS/MAPK signaling pathway associated with rare-human genetic disorders with the goal of developing zebrafish models to study mechanisms of disease. Using the classroom as my laboratory, students in the UAB Science and Technology Honors Program (SciTech) are working alongside me to design and construct CRISPR-guide RNA to target gene sequences in the zebrafish genome homologous to human RAS/MAPK pathway genes. You can check out more details on this course and view the syllabus under the teaching experience tab.
Research in the classroom
2019
Developing zebrafish models of patient-guided rare-neuropsychiatric disorders
Patients with rare-genetic disorders often exhibit epilepsy-like symptoms, such as seizure and developmental delay. This semester in the Science and Technology honors course: Research Approaches: Molecular Genetics, I am working with students to develop loss-of-function (LOF) zebrafish models for patients with predicted LOF or haploinsufficient genetic variants. Using CRISPR-Cas9 gene editing, students are working on teams of 2 to design CRISPR guides targeting genes of interest, screen RNA guide + Cas9 cutting efficiencies in vivo, and analyze early-stage phenotypes in microinjected F0 zebrafish. Successfully mutated zebrafish will be raised, sequenced, and bred to develop LOF zebrafish models of genetic disorders. Students interested in completing their honor's thesis projects using these LOF mutants will then work with me, in collaboration with Dr. Summer Thyme, to identify screenable phenotypes (behavior, brain structure & activity) for which FDA-approved drug libraries can be screened in order to find therapeutic options to potentially treat individual patients.
2018
Inducing mutations in genes involved in the RAS/MAPK signaling pathway to examine disease mechanisms associated with rare rasopathies
The RAS/MAPK signaling pathway is involved in multiple processes associated with development in humans. Mutations in genes of this pathway can cause a break down in cell signaling necessary for proper growth and development, resulting in a spectrum of rare disorders, collectively referred to as rasopathies. I've partnered with Dr. Matt Might, director of UAB's Hugh Kaul Precison Medicine Institute, to target genes in the RAS/MAPK signaling pathway associated with rare-human genetic disorders with the goal of developing zebrafish models to study mechanisms of disease. Using the classroom as my laboratory, students in the UAB Science and Technology Honors Program (SciTech) are working alongside me to design and construct CRISPR-guide RNA to target gene sequences in the zebrafish genome homologous to human RAS/MAPK pathway genes. You can check out more details on this course and view the syllabus under the teaching experience tab.